
09 Feb Using puzzle solutions to aerate alignments
In past blog posts, we have shown snippets of our analysis pipeline and some data samples, and as we are now approaching the step of showing results, we would like to share a peek at the kind of results we are looking for. Obviously, we cannot show everything as most of the results must first be published in peer reviewed scientific journals, but we want to keep leveraging this blog to update you on where we are and provide you an intuitive understanding of what we are doing.
In terms of the big picture, we are starting from genomic sequences and trying to align these sequences with the input of Borderlands 3 players. We started from alignments built by computers, divided these alignments into small chunks, and released these small chunks as puzzles to be solved in the arcade booth within the game. We then grabbed the solutions to these, compared all the player solutions together and extracted the optimal ones, and then all the other solutions that were relatively close to being optimal. This is where we left you at the end of the last blog post.
Since then, we have been using these puzzle results to improve our alignments. Intuitively, the task of sequence alignment is very similar to the Borderlands Science game: the goal is to identify where to insert gaps, or yellow tokens, to align the bricks (nucleotides) as well as possible. As shown on the picture below, we have an initial genomic sequence, and then many puzzle solutions from players for different regions of that sequence that were featured in puzzles, and will influence how we decided to align this sequence.
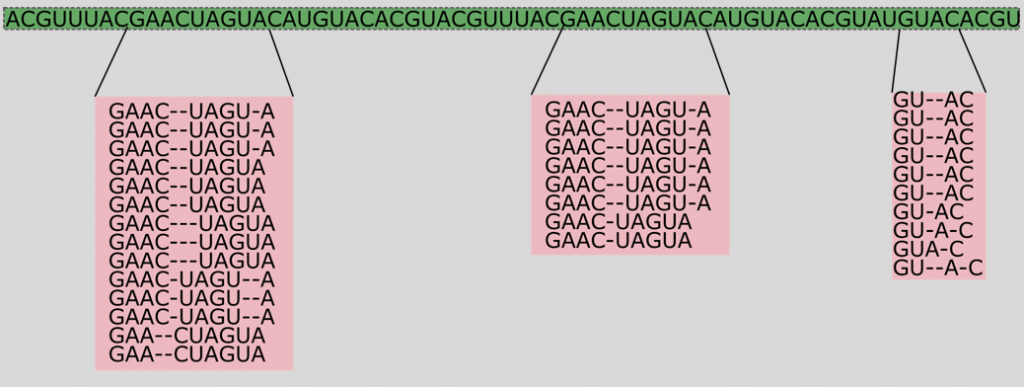
We wanted to show you a before/after picture of a region of an alignment before and after adding this information from the puzzles:

As you can see, the left image includes very few gaps, but many columns in which the identify is not very well defined (many colors are mixed). The right image represents the same data, i.e. the bricks are the same but with much more space between columns, which allows to have less dissimilarity within the same column, and group more nucleotides that belong together.
Alignments are always “messy” to a degree since they include the information that comes from millions of years of evolution, so there will never be a solution where everything is perfectly aligned (as you might have figured out when playing the game!), but the information we get from players appears to have the potential to improve these alignments, which will help us understand the evolution of different species of gut bacteria better, as well as how these evolutionary patterns could influence our health.
While we are reporting about data analysis and obtaining results, please don’t believe the project is over! We are still collecting and regularly uploading new puzzles, and we are custom-adjusting these puzzles to target regions that we don’t know a lot (basically, information we are currently missing), so an individual puzzle solved now can have a very significant impact on our results. In other words, we are using what we know to figure out what we don’t know, and then, with your help, we are filling these gaps in our knowledge.

