17 Aug From Phylo to Borderlands Science
In this post, we shed a light on the origin and conception of Borderlands Science.
Ten year ago, we launched our first citizen science game, Phylo DNA Puzzles, an online casual puzzle game that can be freely played at http://phylo.cs.mcgill.ca. As Borderlands Science, the solutions collected from Phylo help us to analyze DNA sequences and reveal similarities and differences the genomes of various organisms. But there are a couple of key differences.
First, the DNA sequences have different origins. Phylo aims to compute a better mapping of mammalian genes, including human’s genes, associated with a disease. To be more accurate, a region upstream of the genes called the promoter region, which is used to regulate its expression. By contrast, Borderlands Science analyzes microbial DNA sequences sequenced by the Microsetta Initiative. Why is it different? First, Borderlands Science has been designed to compare much more sequences than Phylo. Phylo collects all the gamer’s solutions to build alignments of about one hundred sequences. By contrast, Borderlands Science aims to align of millions of sequences! Next, we know for each sequence in Phylo the species it is coming from. This information allows us to determine prioritize the comparisons of a subset of sequences and score more accurately the solutions. But it is not available in the microbiome data set used by Borderlands Science and we have to resort on a different type of guide (i.e. the tokens you are trying to match at the beginning of each row) to estimate the quality of the puzzle solutions.
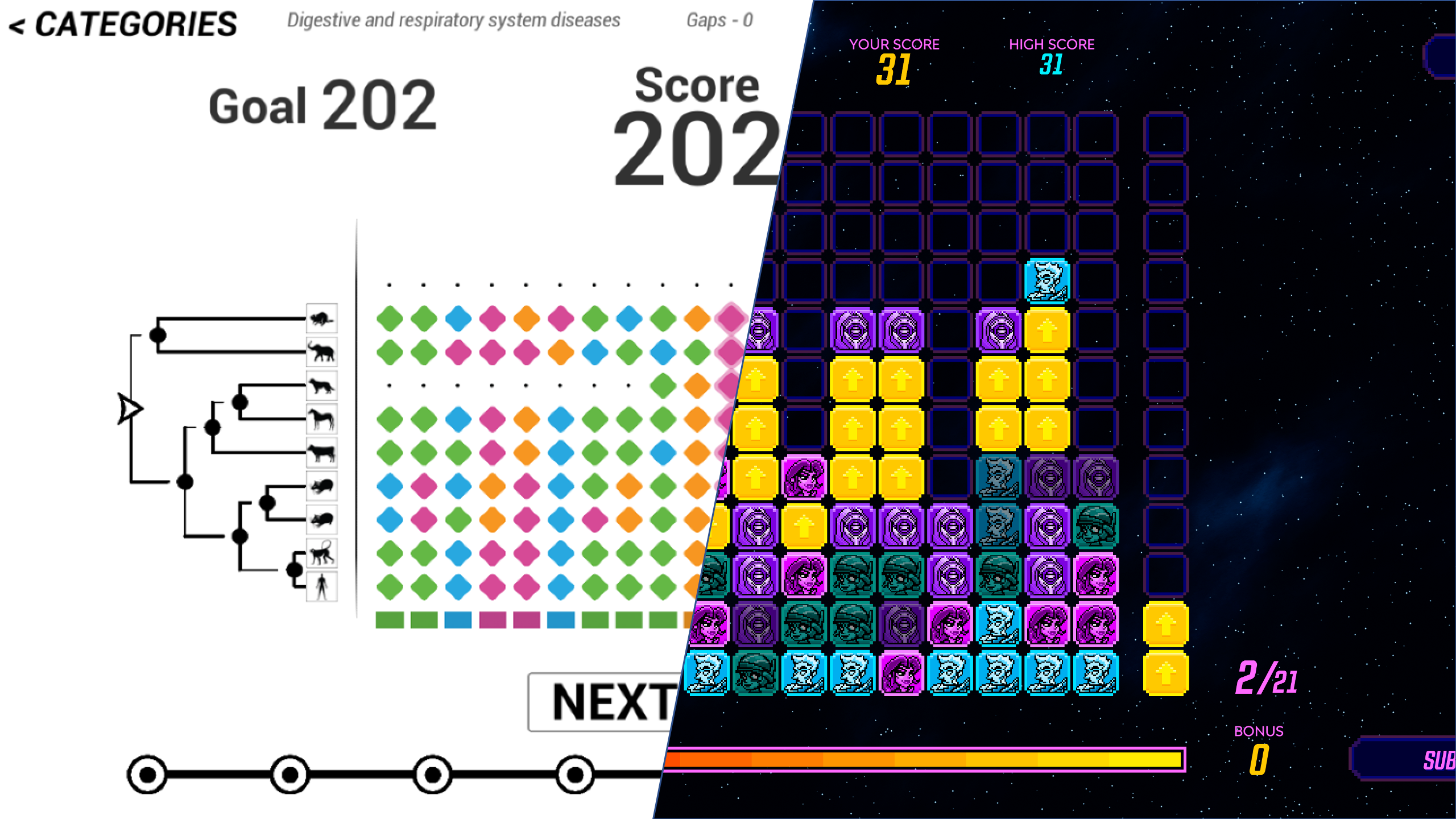
Though, the biggest challenge was probably the design of the game itself. In 2010, one of the main novelties of Phylo was to frame a scientific problem into a casual game that anyone can play with a minimum training and no biology background. Online users can access the game without registration and spend as little as a couple of minutes to play one game and make a contribution. The interface underwent many refactoring though the years, but the basic rules and design element remained the same. You can see an illustration of this interface below (from the 2016 release).
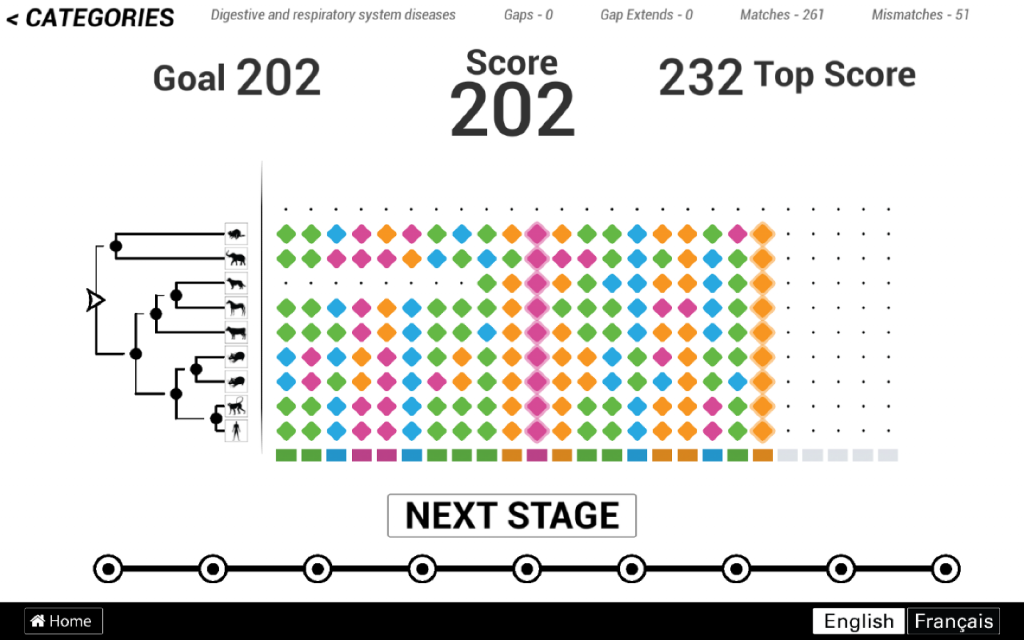
This interface tries to make the most accessible and intelligible presentation of the multiple sequence alignment problem possible. In particular, the scoring function is simplified and the A, C, G, T characters representing the DNA sequences have been replaced by tiles of four different colors. Still, this interface was not as casual as we initially hoped. Did you notice the tree of the left or the ancestral sequence at the bottom? The scoring function too is not easily intelligible for a non-expert.
Anyways, the point is that for Borderlands 3, we needed to bring this concept to a whole different level. We thus sat with the team of Gearbox Studio Québec and worked through the full year of 2019 on bringing Phylo to this:
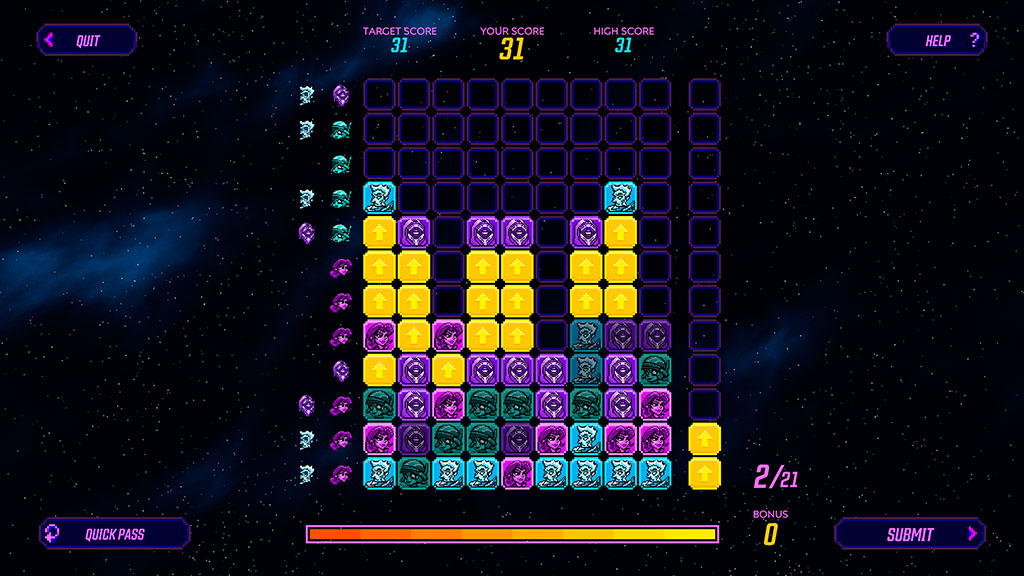
The contrast is striking. The rules are simpler, which makes the game more accessible by a broad audience. No need to mention that the presentation of the task itself is much more fun than in Phylo. We could write a lot about the process of designing Borderlands Science, but instead of that what could be better than a short video directly from the game designer who created Borderlands Science, Gabriel Richard.
To be fair, we should stress that Borderlands Science would not have been possible without the experience accumulated since 2010 with Phylo. We got a lot of feedback and data that helped us to make Borderlands Science more scientifically useful and… more fun! The Phylo project is still running. Recently it took a turn toward more educational uses and we also use it to explore more complex alignments. If you are interested, you can have a look at the following papers.
[1] Phylo: a citizen science approach for improving multiple sequence alignment,
Kawrykow, Alexander, Roumanis, Gary, Kam, Alfred, Kwak, Daniel, Leung, Clarence, Wu, Chu, Zarour, Eleyine, {Phylo players}, Sarmenta, Luis, Blanchette, Mathieu, and Waldispühl, Jérôme
PLoS One, 7, pages e31362, March, 2012.
[2] Open-Phylo: a customizable crowd-computing platform for multiple sequence alignment,
Kwak, Daniel, Kam, Alfred, Becerra, David, Zhou, Qikuan, Hops, Adam, Zarour, Eleyine, Kam, Arthur, Sarmenta, Luis, Blanchette, Mathieu, and Waldispühl, Jérôme
Genome Biol, 14, pages R116, October, 2013.
[3] Crowdsourcing RNA structural alignments with an online computer game,
Waldispühl, Jérôme, Kam, Arthur, and Gardner, Paul P
In Proceedings of the 20th Pacific Symposium on Biocomputing, pages 330-41, 2015.
[4] Lessons from an online massive genomics computer game,
Singh, Akash, Ahsan, Faizy, Blanchette, Mathieu, and Waldispühl, Jérôme
In Proceedings of the Fifth AAAI Conference on Human Computation and Crowdsourcing (HCOMP 2017), 2017.
In the next blog post, we will come back to the analysis of the data we collected with Borderlands Science and perhaps some interesting observations. Stay tuned! It should be fun.

